stats: The R Base Package
R標準の統計解析パッケージ
> library(stats)
バージョン: 3.2.3
| 関数名 | 概略 |
|---|---|
.checkMFClasses |
Functions to Check the Type of Variables passed to Model Frames |
AIC |
Akaike's An Information Criterion |
ARMAacf |
Compute Theoretical ACF for an ARMA Process |
ARMAtoMA |
Convert ARMA Process to Infinite MA Process |
Beta |
The Beta Distribution |
Binomial |
The Binomial Distribution |
Box.test |
Box-Pierce and Ljung-Box Tests |
C |
Sets Contrasts for a Factor |
Cauchy |
The Cauchy Distribution |
Chisquare |
The (non-central) Chi-Squared Distribution |
Distributions |
Distributions in the stats package |
Exponential |
The Exponential Distribution |
FDist |
The F Distribution |
GammaDist |
The Gamma Distribution |
Geometric |
The Geometric Distribution |
HoltWinters |
Holt-Winters Filtering |
Hypergeometric |
The Hypergeometric Distribution |
IQR |
The Interquartile Range |
KalmanLike |
Kalman Filtering |
Logistic |
The Logistic Distribution |
Lognormal |
The Log Normal Distribution |
Multinomial |
The Multinomial Distribution |
NLSstAsymptotic |
Fit the Asymptotic Regression Model |
NLSstClosestX |
Inverse Interpolation |
NLSstLfAsymptote |
Horizontal Asymptote on the Left Side |
NLSstRtAsymptote |
Horizontal Asymptote on the Right Side |
NegBinomial |
The Negative Binomial Distribution |
Normal |
The Normal Distribution |
PP.test |
Phillips-Perron Test for Unit Roots |
Poisson |
The Poisson Distribution |
SSD |
SSD Matrix and Estimated Variance Matrix in Multivariate Models |
SSasymp |
Self-Starting Nls Asymptotic Regression Model |
SSasympOff |
Self-Starting Nls Asymptotic Regression Model with an Offset |
SSasympOrig |
Self-Starting Nls Asymptotic Regression Model through the Origin |
SSbiexp |
Self-Starting Nls Biexponential model |
SSfol |
Self-Starting Nls First-order Compartment Model |
SSfpl |
Self-Starting Nls Four-Parameter Logistic Model |
SSgompertz |
Self-Starting Nls Gompertz Growth Model |
SSlogis |
Self-Starting Nls Logistic Model |
SSmicmen |
Self-Starting Nls Michaelis-Menten Model |
SSweibull |
Self-Starting Nls Weibull Growth Curve Model |
SignRank |
Distribution of the Wilcoxon Signed Rank Statistic |
StructTS |
Fit Structural Time Series |
TDist |
The Student t Distribution |
Tukey |
The Studentized Range Distribution |
TukeyHSD |
Compute Tukey Honest Significant Differences |
Uniform |
The Uniform Distribution |
Weibull |
The Weibull Distribution |
Wilcoxon |
Distribution of the Wilcoxon Rank Sum Statistic |
acf |
Auto- and Cross- Covariance and -Correlation Function Estimation |
acf2AR |
Compute an AR Process Exactly Fitting an ACF |
add1 |
Add or Drop All Possible Single Terms to a Model |
addmargins |
Puts Arbitrary Margins on Multidimensional Tables or Arrays |
aggregate |
Compute Summary Statistics of Data Subsets |
alias |
Find Aliases (Dependencies) in a Model |
anova |
Anova Tables |
anova.glm |
Analysis of Deviance for Generalized Linear Model Fits |
anova.lm |
ANOVA for Linear Model Fits |
anova.mlm |
Comparisons between Multivariate Linear Models |
ansari.test |
Ansari-Bradley Test |
aov |
Fit an Analysis of Variance Model |
approxfun |
Interpolation Functions |
ar |
Fit Autoregressive Models to Time Series |
ar.ols |
Fit Autoregressive Models to Time Series by OLS |
arima |
ARIMA Modelling of Time Series |
arima.sim |
Simulate from an ARIMA Model |
arima0 |
ARIMA Modelling of Time Series - Preliminary Version |
as.hclust |
Convert Objects to Class hclust |
asOneSidedFormula |
Convert to One-Sided Formula |
ave |
Group Averages Over Level Combinations of Factors |
bartlett.test |
Bartlett Test of Homogeneity of Variances |
binom.test |
Exact Binomial Test |
biplot |
Biplot of Multivariate Data |
biplot.princomp |
Biplot for Principal Components |
bw.nrd0 |
Bandwidth Selectors for Kernel Density Estimation |
cancor |
Canonical Correlations |
case.names |
Case and Variable Names of Fitted Models |
chisq.test |
Pearson's Chi-squared Test for Count Data |
cmdscale |
Classical (Metric) Multidimensional Scaling |
coef |
Extract Model Coefficients |
complete.cases |
Find Complete Cases |
confint |
Confidence Intervals for Model Parameters |
constrOptim |
Linearly Constrained Optimization |
contr.helmert |
(Possibly Sparse) Contrast Matrices |
contrasts |
Get and Set Contrast Matrices |
convolve |
Convolution of Sequences via FFT |
cophenetic |
Cophenetic Distances for a Hierarchical Clustering |
cor |
Correlation, Variance and Covariance (Matrices) |
cor.test |
Test for Association/Correlation Between Paired Samples |
cov.wt |
Weighted Covariance Matrices |
cpgram |
Plot Cumulative Periodogram |
cutree |
Cut a Tree into Groups of Data |
decompose |
Classical Seasonal Decomposition by Moving Averages |
delete.response |
Modify Terms Objects |
dendrapply |
Apply a Function to All Nodes of a Dendrogram |
dendrogram |
General Tree Structures |
density |
Kernel Density Estimation |
deriv |
Symbolic and Algorithmic Derivatives of Simple Expressions |
deviance |
Model Deviance |
df.residual |
Residual Degrees-of-Freedom |
diff.ts |
Methods for Time Series Objects |
diffinv |
Discrete Integration: Inverse of Differencing |
dist |
Distance Matrix Computation |
dummy.coef |
Extract Coefficients in Original Coding |
ecdf |
Empirical Cumulative Distribution Function |
eff.aovlist |
Compute Efficiencies of Multistratum Analysis of Variance |
effects |
Effects from Fitted Model |
embed |
Embedding a Time Series |
expand.model.frame |
Add new variables to a model frame |
extractAIC |
Extract AIC from a Fitted Model |
factanal |
Factor Analysis |
factor.scope |
Compute Allowed Changes in Adding to or Dropping from a Formula |
family |
Family Objects for Models |
family.glm |
Accessing Generalized Linear Model Fits |
family.lm |
Accessing Linear Model Fits |
fft |
Fast Discrete Fourier Transform |
filter |
Linear Filtering on a Time Series |
fisher.test |
Fisher's Exact Test for Count Data |
fitted |
Extract Model Fitted Values |
fivenum |
Tukey Five-Number Summaries |
fligner.test |
Fligner-Killeen Test of Homogeneity of Variances |
formula |
Model Formulae |
formula.nls |
Extract Model Formula from nls Object |
friedman.test |
Friedman Rank Sum Test |
ftable |
Flat Contingency Tables |
ftable.formula |
Formula Notation for Flat Contingency Tables |
getInitial |
Get Initial Parameter Estimates |
glm |
Fitting Generalized Linear Models |
glm.control |
Auxiliary for Controlling GLM Fitting |
hclust |
Hierarchical Clustering |
heatmap |
Draw a Heat Map |
identify.hclust |
Identify Clusters in a Dendrogram |
influence.measures |
Regression Deletion Diagnostics |
integrate |
Integration of One-Dimensional Functions |
interaction.plot |
Two-way Interaction Plot |
is.empty.model |
Test if a Model's Formula is Empty |
isoreg |
Isotonic / Monotone Regression |
kernapply |
Apply Smoothing Kernel |
kernel |
Smoothing Kernel Objects |
kmeans |
K-Means Clustering |
kruskal.test |
Kruskal-Wallis Rank Sum Test |
ks.test |
Kolmogorov-Smirnov Tests |
ksmooth |
Kernel Regression Smoother |
lag |
Lag a Time Series |
lag.plot |
Time Series Lag Plots |
line |
Robust Line Fitting |
listof |
A Class for Lists of (Parts of) Model Fits |
lm |
Fitting Linear Models |
lm.fit |
Fitter Functions for Linear Models |
lm.influence |
Regression Diagnostics |
loadings |
Print Loadings in Factor Analysis |
loess |
Local Polynomial Regression Fitting |
loess.control |
Set Parameters for Loess |
logLik |
Extract Log-Likelihood |
loglin |
Fitting Log-Linear Models |
lowess |
Scatter Plot Smoothing |
ls.diag |
Compute Diagnostics for 'lsfit' Regression Results |
ls.print |
Print 'lsfit' Regression Results |
lsfit |
Find the Least Squares Fit |
mad |
Median Absolute Deviation |
mahalanobis |
Mahalanobis Distance |
make.link |
Create a Link for GLM Families |
makepredictcall |
Utility Function for Safe Prediction |
manova |
Multivariate Analysis of Variance |
mantelhaen.test |
Cochran-Mantel-Haenszel Chi-Squared Test for Count Data |
mauchly.test |
Mauchly's Test of Sphericity |
mcnemar.test |
McNemar's Chi-squared Test for Count Data |
median |
Median Value |
medpolish |
Median Polish of a Matrix |
model.extract |
Extract Components from a Model Frame |
model.frame |
Extracting the Model Frame from a Formula or Fit |
model.matrix |
Construct Design Matrices |
model.tables |
Compute Tables of Results from an Aov Model Fit |
monthplot |
Plot a Seasonal or other Subseries from a Time Series |
mood.test |
Mood Two-Sample Test of Scale |
na.action |
NA Action |
na.contiguous |
Find Longest Contiguous Stretch of non-NAs |
na.fail |
Handle Missing Values in Objects |
naprint |
Adjust for Missing Values |
naresid |
Adjust for Missing Values |
nextn |
Highly Composite Numbers |
nlm |
Non-Linear Minimization |
nlminb |
Optimization using PORT routines |
nls |
Nonlinear Least Squares |
nls.control |
Control the Iterations in nls |
nobs |
Extract the Number of Observations from a Fit. |
numericDeriv |
Evaluate Derivatives Numerically |
offset |
Include an Offset in a Model Formula |
oneway.test |
Test for Equal Means in a One-Way Layout |
optim |
General-purpose Optimization |
optimize |
One Dimensional Optimization |
order.dendrogram |
Ordering or Labels of the Leaves in a Dendrogram |
p.adjust |
Adjust P-values for Multiple Comparisons |
pairwise.prop.test |
Pairwise comparisons for proportions |
pairwise.t.test |
Pairwise t tests |
pairwise.table |
Tabulate p values for pairwise comparisons |
pairwise.wilcox.test |
Pairwise Wilcoxon Rank Sum Tests |
plot.HoltWinters |
Plot function for HoltWinters objects |
plot.acf |
Plot Autocovariance and Autocorrelation Functions |
plot.density |
Plot Method for Kernel Density Estimation |
plot.isoreg |
Plot Method for isoreg Objects |
plot.lm |
Plot Diagnostics for an lm Object |
plot.ppr |
Plot Ridge Functions for Projection Pursuit Regression Fit |
plot.profile.nls |
Plot a profile.nls Object |
plot.spec |
Plotting Spectral Densities |
plot.stepfun |
Plot Step Functions |
plot.stl |
Methods for STL Objects |
plot.ts |
Plotting Time-Series Objects |
poisson.test |
Exact Poisson tests |
poly |
Compute Orthogonal Polynomials |
power |
Create a Power Link Object |
power.anova.test |
Power Calculations for Balanced One-Way Analysis of Variance Tests |
power.prop.test |
Power Calculations for Two-Sample Test for Proportions |
power.t.test |
Power calculations for one and two sample t tests |
ppoints |
Ordinates for Probability Plotting |
ppr |
Projection Pursuit Regression |
prcomp |
Principal Components Analysis |
predict |
Model Predictions |
predict.Arima |
Forecast from ARIMA fits |
predict.HoltWinters |
Prediction Function for Fitted Holt-Winters Models |
predict.glm |
Predict Method for GLM Fits |
predict.lm |
Predict method for Linear Model Fits |
predict.loess |
Predict Loess Curve or Surface |
predict.nls |
Predicting from Nonlinear Least Squares Fits |
predict.smooth.spline |
Predict from Smoothing Spline Fit |
preplot |
Pre-computations for a Plotting Object |
princomp |
Principal Components Analysis |
print.power.htest |
Print method for power calculation object |
print.ts |
Printing and Formatting of Time-Series Objects |
printCoefmat |
Print Coefficient Matrices |
profile |
Generic Function for Profiling Models |
profile.nls |
Method for Profiling nls Objects |
proj |
Projections of Models |
prop.test |
Test of Equal or Given Proportions |
prop.trend.test |
Test for trend in proportions |
qbirthday |
Probability of coincidences |
qqnorm |
Quantile-Quantile Plots |
quade.test |
Quade Test |
quantile |
Sample Quantiles |
r2dtable |
Random 2-way Tables with Given Marginals |
rWishart |
Random Wishart Distributed Matrices |
read.ftable |
Manipulate Flat Contingency Tables |
rect.hclust |
Draw Rectangles Around Hierarchical Clusters |
relevel |
Reorder Levels of Factor |
reorder.default |
Reorder Levels of a Factor |
reorder.dendrogram |
Reorder a Dendrogram |
replications |
Number of Replications of Terms |
reshape |
Reshape Grouped Data |
residuals |
Extract Model Residuals |
runmed |
Running Medians - Robust Scatter Plot Smoothing |
scatter.smooth |
Scatter Plot with Smooth Curve Fitted by Loess |
screeplot |
Screeplots |
sd |
Standard Deviation |
se.contrast |
Standard Errors for Contrasts in Model Terms |
selfStart |
Construct Self-starting Nonlinear Models |
setNames |
Set the Names in an Object |
shapiro.test |
Shapiro-Wilk Normality Test |
simulate |
Simulate Responses |
smooth |
Tukey's (Running Median) Smoothing |
smooth.spline |
Fit a Smoothing Spline |
smoothEnds |
End Points Smoothing (for Running Medians) |
sortedXyData |
Create a 'sortedXyData' Object |
spec.ar |
Estimate Spectral Density of a Time Series from AR Fit |
spec.pgram |
Estimate Spectral Density of a Time Series by a Smoothed Periodogram |
spec.taper |
Taper a Time Series by a Cosine Bell |
spectrum |
Spectral Density Estimation |
splinefun |
Interpolating Splines |
start |
Encode the Terminal Times of Time Series |
stat.anova |
GLM Anova Statistics |
stats-deprecated |
Deprecated Functions in Package 'stats' |
stats-package |
The R Stats Package |
step |
Choose a model by AIC in a Stepwise Algorithm |
stepfun |
Step Functions - Creation and Class |
stl |
Seasonal Decomposition of Time Series by Loess |
summary.aov |
Summarize an Analysis of Variance Model |
summary.glm |
Summarizing Generalized Linear Model Fits |
summary.lm |
Summarizing Linear Model Fits |
summary.manova |
Summary Method for Multivariate Analysis of Variance |
summary.nls |
Summarizing Non-Linear Least-Squares Model Fits |
summary.princomp |
Summary method for Principal Components Analysis |
supsmu |
Friedman's SuperSmoother |
symnum |
Symbolic Number Coding |
t.test |
Student's t-Test |
termplot |
Plot Regression Terms |
terms |
Model Terms |
terms.formula |
Construct a terms Object from a Formula |
terms.object |
Description of Terms Objects |
time |
Sampling Times of Time Series |
toeplitz |
Form Symmetric Toeplitz Matrix |
ts |
Time-Series Objects |
ts.plot |
Plot Multiple Time Series |
ts.union |
Bind Two or More Time Series |
tsSmooth |
Use Fixed-Interval Smoothing on Time Series |
tsdiag |
Diagnostic Plots for Time-Series Fits |
tsp |
Tsp Attribute of Time-Series-like Objects |
uniroot |
One Dimensional Root (Zero) Finding |
update |
Update and Re-fit a Model Call |
update.formula |
Model Updating |
var.test |
F Test to Compare Two Variances |
varimax |
Rotation Methods for Factor Analysis |
vcov |
Calculate Variance-Covariance Matrix for a Fitted Model Object |
weighted.mean |
Weighted Arithmetic Mean |
weighted.residuals |
Compute Weighted Residuals |
weights |
Extract Model Weights |
wilcox.test |
Wilcoxon Rank Sum and Signed Rank Tests |
window |
Time Windows |
xtabs |
Cross Tabulation |
Distributions
確率分布
> # dbeta
> # dbinom
> # dcauchy
> # ...
Lognormal / dlnorm / plnorm / qlnorm / rlnorm
対数正規分布
Arguments
- x, q
- p
- meanlog, sdlog
- log, log.p
- lower.tail
> rlnorm(n = 10)
[1] 4.1484730 2.3108145 0.3768001 0.1954081 0.4252516 0.2927458 1.2176975
[8] 0.1893988 1.3688176 0.4578168
acf / pacf / ccf
自己相関のプロット
> acf(lh)
> acf(lh, type = "covariance")
> pacf(lh)
aggregate
要約統計量の算出
> iris %>% aggregate(. ~ Species, data = ., mean)
Species Sepal.Length Sepal.Width Petal.Length Petal.Width
1 setosa 5.006 3.428 1.462 0.246
2 versicolor 5.936 2.770 4.260 1.326
3 virginica 6.588 2.974 5.552 2.026
ave
> data(warpbreaks)
> ave(warpbreaks$breaks, warpbreaks$wool)
[1] 31.03704 31.03704 31.03704 31.03704 31.03704 31.03704 31.03704
[8] 31.03704 31.03704 31.03704 31.03704 31.03704 31.03704 31.03704
[15] 31.03704 31.03704 31.03704 31.03704 31.03704 31.03704 31.03704
[22] 31.03704 31.03704 31.03704 31.03704 31.03704 31.03704 25.25926
[29] 25.25926 25.25926 25.25926 25.25926 25.25926 25.25926 25.25926
[36] 25.25926 25.25926 25.25926 25.25926 25.25926 25.25926 25.25926
[43] 25.25926 25.25926 25.25926 25.25926 25.25926 25.25926 25.25926
[50] 25.25926 25.25926 25.25926 25.25926 25.25926
> iris %$% ave(Sepal.Length, Species)
[1] 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006
[12] 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006
[23] 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006
[34] 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006 5.006
[45] 5.006 5.006 5.006 5.006 5.006 5.006 5.936 5.936 5.936 5.936 5.936
[56] 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936
[67] 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936
[78] 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936
[89] 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936 5.936
[100] 5.936 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588
[111] 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588
[122] 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588
[133] 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588 6.588
[144] 6.588 6.588 6.588 6.588 6.588 6.588 6.588
complete.cases
欠損値の確認
> complete.cases(c(1, 2, NA, 4))
[1] TRUE TRUE FALSE TRUE
decompose
> x <- c(-50, 175, 149, 214, 247, 237, 225, 329, 729, 809,
+ 530, 489, 540, 457, 195, 176, 337, 239, 128, 102, 232, 429, 3,
+ 98, 43, -141, -77, -13, 125, 361, -45, 184) %>%
+ ts(start = c(1951, 1), end = c(1958, 4), frequency = 4)
> decompose(x)
$x
Qtr1 Qtr2 Qtr3 Qtr4
1951 -50 175 149 214
1952 247 237 225 329
1953 729 809 530 489
1954 540 457 195 176
1955 337 239 128 102
1956 232 429 3 98
1957 43 -141 -77 -13
1958 125 361 -45 184
$seasonal
Qtr1 Qtr2 Qtr3 Qtr4
1951 62.45982 86.17411 -88.37946 -60.25446
1952 62.45982 86.17411 -88.37946 -60.25446
1953 62.45982 86.17411 -88.37946 -60.25446
1954 62.45982 86.17411 -88.37946 -60.25446
1955 62.45982 86.17411 -88.37946 -60.25446
1956 62.45982 86.17411 -88.37946 -60.25446
1957 62.45982 86.17411 -88.37946 -60.25446
1958 62.45982 86.17411 -88.37946 -60.25446
$trend
Qtr1 Qtr2 Qtr3 Qtr4
1951 NA NA 159.125 204.000
1952 221.250 245.125 319.750 451.500
1953 561.125 619.250 615.625 548.000
1954 462.125 381.125 316.625 264.000
1955 228.375 210.750 188.375 199.000
1956 207.125 191.000 166.875 72.000
1957 -9.250 -33.125 -36.750 36.250
1958 103.000 131.625 NA NA
$random
Qtr1 Qtr2 Qtr3 Qtr4
1951 NA NA 78.254464 70.254464
1952 -36.709821 -94.299107 -6.370536 -62.245536
1953 105.415179 103.575893 2.754464 1.254464
1954 15.415179 -10.299107 -33.245536 -27.745536
1955 46.165179 -57.924107 28.004464 -36.745536
1956 -37.584821 151.825893 -75.495536 86.254464
1957 -10.209821 -194.049107 48.129464 11.004464
1958 -40.459821 143.200893 NA NA
$figure
[1] 62.45982 86.17411 -88.37946 -60.25446
$type
[1] "additive"
attr(,"class")
[1] "decomposed.ts"
dist
距離行列
Arguments
- x
- method...
euclidean,maximum,manhattan,canberra,binary,minkowski - diag... 出力オプション
- upper... 出力オプション
- p
- m
- digits, justify
- right
- ...
> x <- matrix(rnorm(100), nrow = 5)
> dist(x)
1 2 3 4
2 5.535243
3 6.339792 7.941767
4 7.089858 7.070399 4.346966
5 5.811479 7.041772 5.518783 6.457818
> dist(x, diag = TRUE)
1 2 3 4 5
1 0.000000
2 5.535243 0.000000
3 6.339792 7.941767 0.000000
4 7.089858 7.070399 4.346966 0.000000
5 5.811479 7.041772 5.518783 6.457818 0.000000
> dist(x, upper = TRUE)
1 2 3 4 5
1 5.535243 6.339792 7.089858 5.811479
2 5.535243 7.941767 7.070399 7.041772
3 6.339792 7.941767 4.346966 5.518783
4 7.089858 7.070399 4.346966 6.457818
5 5.811479 7.041772 5.518783 6.457818
family
モデリングのための確率分布オブジェクト
Arguments
- link
- variance
- object
- ...
filter
時系列データのフィルタリング
Arguments
- x
- filter
- method
- sides
- circular
> x <- 1:100
> filter(x, rep(1, 3))
Time Series:
Start = 1
End = 100
Frequency = 1
[1] NA 6 9 12 15 18 21 24 27 30 33 36 39 42 45 48 51
[18] 54 57 60 63 66 69 72 75 78 81 84 87 90 93 96 99 102
[35] 105 108 111 114 117 120 123 126 129 132 135 138 141 144 147 150 153
[52] 156 159 162 165 168 171 174 177 180 183 186 189 192 195 198 201 204
[69] 207 210 213 216 219 222 225 228 231 234 237 240 243 246 249 252 255
[86] 258 261 264 267 270 273 276 279 282 285 288 291 294 297 NA
> filter(x, rep(1, 3), sides = 1, circular = TRUE)
Time Series:
Start = 1
End = 100
Frequency = 1
[1] 200 103 6 9 12 15 18 21 24 27 30 33 36 39 42 45 48
[18] 51 54 57 60 63 66 69 72 75 78 81 84 87 90 93 96 99
[35] 102 105 108 111 114 117 120 123 126 129 132 135 138 141 144 147 150
[52] 153 156 159 162 165 168 171 174 177 180 183 186 189 192 195 198 201
[69] 204 207 210 213 216 219 222 225 228 231 234 237 240 243 246 249 252
[86] 255 258 261 264 267 270 273 276 279 282 285 288 291 294 297
hclust
階層クラスタリング
Arguments
- d... distクラスオブジェクト
- method...
ward.D,ward.D2,single,complete,average,mcquitty,median,centroid - members
- x
- hang
- check
- labels
- axes, frame.plot, ann
- main, sub, xlab, ylab
- ...
> dist(USArrests) %>% hclust(method = "ave")
Call:
hclust(d = ., method = "ave")
Cluster method : average
Distance : euclidean
Number of objects: 50
lag
時系列の差分
Arguments
- x
- k
- ...
> lag(x = ldeaths, k = 12)
Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
1973 3035 2552 2704 2554 2014 1655 1721 1524 1596 2074 2199 2512
1974 2933 2889 2938 2497 1870 1726 1607 1545 1396 1787 2076 2837
1975 2787 3891 3179 2011 1636 1580 1489 1300 1356 1653 2013 2823
1976 3102 2294 2385 2444 1748 1554 1498 1361 1346 1564 1640 2293
1977 2815 3137 2679 1969 1870 1633 1529 1366 1357 1570 1535 2491
1978 3084 2605 2573 2143 1693 1504 1461 1354 1333 1492 1781 1915
margin.table
> matrix(1:4, 2) %>% {
+ print(margin.table(., 1))
+ margin.table(., 2)
+ }
[1] 4 6
[1] 3 7
na.fail / na.omit / na.exclude / na.pass
オブジェクト内の欠損処理
> DT <- data.frame(x = c(1, 2, 3), y = c(0, 10, NA))
> DT %>% {
+ print(na.omit(.))
+ print(na.pass(.))
+ }
x y
1 1 0
2 2 10
x y
1 1 0
2 2 10
3 3 NA
p.adjust
P値の補正
> set.seed(123)
> x <- rnorm(50, mean = c(rep(0, 25), rep(3, 25)))
> p <- 2*pnorm(sort(-abs(x)))
>
> round(p, 3)
[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.001
[12] 0.002 0.003 0.004 0.005 0.007 0.007 0.009 0.009 0.011 0.021 0.049
[23] 0.061 0.063 0.074 0.083 0.086 0.119 0.189 0.206 0.221 0.286 0.305
[34] 0.466 0.483 0.492 0.532 0.575 0.578 0.619 0.636 0.645 0.656 0.689
[45] 0.719 0.818 0.827 0.897 0.912 0.944
> round(p.adjust(p), 3)
[1] 0.000 0.001 0.001 0.005 0.005 0.006 0.006 0.007 0.009 0.016 0.024
[12] 0.063 0.125 0.131 0.189 0.239 0.240 0.291 0.301 0.350 0.635 1.000
[23] 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
[34] 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
[45] 1.000 1.000 1.000 1.000 1.000 1.000
> round(p.adjust(p, "BH"), 3)
[1] 0.000 0.000 0.000 0.001 0.001 0.001 0.001 0.001 0.001 0.002 0.003
[12] 0.007 0.013 0.013 0.017 0.021 0.021 0.024 0.025 0.028 0.050 0.112
[23] 0.130 0.130 0.148 0.159 0.160 0.213 0.326 0.343 0.356 0.446 0.462
[34] 0.684 0.684 0.684 0.719 0.741 0.741 0.763 0.763 0.763 0.763 0.782
[45] 0.799 0.880 0.880 0.930 0.930 0.944
procomp
主成分分析を実施
Arguments
- formula
- data
- subset
- na.action
- ...
- x
- retx
- center... logical. 中央化(中心移動)の有無
- scale... logical. 標準化の有無
- object
- newdata
spectrum
> spectrum(lh)
start / end
時系列データの始点と終点
> xts::xts(rnorm(21), as.Date((Sys.Date() %>% as.numeric() - 20):Sys.Date() %>% as.numeric(), origin = "1970-01-01")) %>% {
+ start(.) %>% print()
+ end(.)
+ }
[1] "2016-02-16"
[1] "2016-03-07"
> data("presidents")
> presidents %>% {
+ start(.) %>% print()
+ end(.)
+ }
[1] 1945 1
[1] 1974 4
time / cycle/ deltat / frequency
時系列データに関する処理諸々
> time(presidents) %>% as.vector()
[1] 1945.00 1945.25 1945.50 1945.75 1946.00 1946.25 1946.50 1946.75
[9] 1947.00 1947.25 1947.50 1947.75 1948.00 1948.25 1948.50 1948.75
[17] 1949.00 1949.25 1949.50 1949.75 1950.00 1950.25 1950.50 1950.75
[25] 1951.00 1951.25 1951.50 1951.75 1952.00 1952.25 1952.50 1952.75
[33] 1953.00 1953.25 1953.50 1953.75 1954.00 1954.25 1954.50 1954.75
[41] 1955.00 1955.25 1955.50 1955.75 1956.00 1956.25 1956.50 1956.75
[49] 1957.00 1957.25 1957.50 1957.75 1958.00 1958.25 1958.50 1958.75
[57] 1959.00 1959.25 1959.50 1959.75 1960.00 1960.25 1960.50 1960.75
[65] 1961.00 1961.25 1961.50 1961.75 1962.00 1962.25 1962.50 1962.75
[73] 1963.00 1963.25 1963.50 1963.75 1964.00 1964.25 1964.50 1964.75
[81] 1965.00 1965.25 1965.50 1965.75 1966.00 1966.25 1966.50 1966.75
[89] 1967.00 1967.25 1967.50 1967.75 1968.00 1968.25 1968.50 1968.75
[97] 1969.00 1969.25 1969.50 1969.75 1970.00 1970.25 1970.50 1970.75
[105] 1971.00 1971.25 1971.50 1971.75 1972.00 1972.25 1972.50 1972.75
[113] 1973.00 1973.25 1973.50 1973.75 1974.00 1974.25 1974.50 1974.75
> cycle(presidents)
Qtr1 Qtr2 Qtr3 Qtr4
1945 1 2 3 4
1946 1 2 3 4
1947 1 2 3 4
1948 1 2 3 4
1949 1 2 3 4
1950 1 2 3 4
1951 1 2 3 4
1952 1 2 3 4
1953 1 2 3 4
1954 1 2 3 4
1955 1 2 3 4
1956 1 2 3 4
1957 1 2 3 4
1958 1 2 3 4
1959 1 2 3 4
1960 1 2 3 4
1961 1 2 3 4
1962 1 2 3 4
1963 1 2 3 4
1964 1 2 3 4
1965 1 2 3 4
1966 1 2 3 4
1967 1 2 3 4
1968 1 2 3 4
1969 1 2 3 4
1970 1 2 3 4
1971 1 2 3 4
1972 1 2 3 4
1973 1 2 3 4
1974 1 2 3 4
> deltat(presidents)
[1] 0.25
> frequency(presidents)
[1] 4
ts
時系列オブジェクト
Arguments
- data
- start
- end
- frequency
- deltat
- ts.eps
- class
- names
- x
- ...
> ts(1:10, frequency = 4, start = c(1959, 2))
Qtr1 Qtr2 Qtr3 Qtr4
1959 1 2 3
1960 4 5 6 7
1961 8 9 10
Value
- sdev... 主成分の標準偏差
- rotation... 主成分負荷量(各変数と主成分との相関係数)
- x
- center, scale
> library(vegan)
Loading required package: permute
Attaching package: 'permute'
The following object is masked from 'package:devtools':
check
Loading required package: lattice
This is vegan 2.3-4
> data(dune)
> tmp <- prcomp(dune, scale = TRUE)
> tmp
Standard deviations:
[1] 2.651876e+00 2.235468e+00 1.885408e+00 1.626053e+00 1.462502e+00
[6] 1.325825e+00 1.215869e+00 1.147346e+00 1.052556e+00 8.994324e-01
[11] 8.632887e-01 8.346228e-01 7.582115e-01 5.982967e-01 4.717356e-01
[16] 4.687690e-01 3.882168e-01 3.631904e-01 2.519873e-01 2.196148e-16
Rotation:
PC1 PC2 PC3 PC4 PC5
Achimill 0.277358149 -0.015906183 -0.171540078 -0.049173592 -0.29797708
Agrostol -0.270446458 0.225338171 0.075149364 -0.022996313 0.02558700
Airaprae 0.001618847 -0.334422632 0.274278753 0.056419555 -0.21157505
Alopgeni -0.099368966 0.262715304 0.202867708 0.183634250 0.07939982
Anthodor 0.202286085 -0.252994352 -0.025027518 0.149016455 -0.25727867
Bellpere 0.201572477 0.127576066 0.038861550 -0.322454771 -0.07154160
Bromhord 0.225274286 0.118823565 -0.002994863 -0.251045921 -0.22298470
Chenalbu -0.054616149 0.119088370 0.135179981 0.232444003 -0.12138149
Cirsarve 0.016649960 0.149380662 0.193439742 -0.189189359 0.05836970
Comapalu -0.161477760 -0.039960081 -0.187024484 -0.106012285 -0.19183468
Eleopalu -0.293597121 -0.006473954 -0.227685384 -0.094858485 -0.04635077
Elymrepe 0.109724080 0.240308038 0.133618192 -0.093621154 -0.08257644
Empenigr -0.006634467 -0.306810069 0.291414519 0.041757498 -0.08967878
Hyporadi 0.017519532 -0.352770478 0.288137299 0.004269798 -0.03521201
Juncarti -0.238810633 0.054362038 -0.104288726 0.005407353 0.12006764
Juncbufo -0.030874269 0.168783055 0.160848821 0.394627384 0.05976745
Lolipere 0.260781049 0.126992599 -0.053228671 -0.173046076 0.19908008
Planlanc 0.260637241 -0.115975258 -0.252281165 0.152008787 0.12508002
Poaprat 0.247586440 0.211405188 0.045967251 -0.147978053 0.18606513
Poatriv 0.147625073 0.320567091 0.098901591 0.195193384 -0.13875194
Ranuflam -0.306240477 0.007882711 -0.168936684 -0.028939692 -0.05685098
Rumeacet 0.177246450 0.004371686 -0.205222489 0.406962363 0.05580819
Sagiproc -0.037114076 0.070055086 0.402043149 0.083191067 0.18822893
Salirepe -0.115885992 -0.237949108 0.034692521 -0.100795686 0.16147267
Scorautu 0.170911118 -0.232081941 0.135559337 -0.107874951 0.16122343
Trifprat 0.179727680 -0.045658643 -0.260817422 0.328898976 0.06476554
Trifrepe 0.167398084 0.019578344 -0.113457105 -0.022272789 -0.08044923
Vicilath 0.112386371 -0.106027466 -0.028724352 -0.238177831 0.38835725
Bracruta -0.025688498 -0.141949026 -0.141161217 0.090465605 0.51332296
Callcusp -0.244915366 -0.037979317 -0.222350635 -0.113087021 -0.13890022
PC6 PC7 PC8 PC9 PC10
Achimill -0.018250222 -0.006282728 -0.1465146630 -0.037072033 -0.284943943
Agrostol 0.186756604 0.245082868 0.0123098484 -0.119510658 -0.110920492
Airaprae 0.096422424 -0.097712511 0.0321399888 -0.009575434 -0.099636156
Alopgeni -0.105946895 0.020135820 -0.0824459661 0.190755977 -0.332376870
Anthodor 0.156125796 0.026640693 -0.0500822271 -0.115352982 -0.235971913
Bellpere 0.085903724 0.187617173 -0.2832554823 0.139865201 0.126977572
Bromhord 0.099278861 0.281830918 -0.1759418372 -0.012395382 -0.215272689
Chenalbu -0.354866757 0.111081984 -0.2457657313 -0.442523896 0.230253188
Cirsarve 0.418278378 0.321366302 0.1896615933 -0.317577712 0.082926161
Comapalu -0.140848501 0.213691628 0.4385138707 0.162039456 0.272304254
Eleopalu 0.086146760 0.039331789 -0.0006513118 0.039218528 -0.335830173
Elymrepe 0.262309219 -0.245075057 0.0238913356 0.339835808 0.352362019
Empenigr 0.093431268 0.077015555 -0.0416727213 0.185013217 -0.019670023
Hyporadi -0.004373535 -0.049490532 0.0919817016 -0.001399784 -0.103591182
Juncarti 0.154461344 -0.179380327 -0.2079525280 0.271134136 -0.283072280
Juncbufo -0.201014256 0.039793136 -0.0083079686 0.182621410 0.031942060
Lolipere 0.018960837 -0.180667416 0.1327932031 -0.077020674 -0.113576629
Planlanc 0.044506375 0.056964592 -0.0185270317 -0.214586350 0.016949100
Poaprat -0.033908359 -0.229590251 -0.0138181427 -0.004776205 -0.023703719
Poatriv -0.028641057 0.103937114 -0.2547465226 0.011586367 -0.038789045
Ranuflam -0.042350839 0.090057291 -0.2759871539 -0.125750626 0.001195093
Rumeacet 0.246487606 0.113785393 0.0366406873 0.134230105 0.114249032
Sagiproc 0.115864362 0.311970978 0.1808234119 -0.042954530 -0.140063426
Salirepe 0.118664904 0.045789547 -0.5063249251 0.042911714 0.335199480
Scorautu -0.173373768 0.251032453 -0.1852370018 0.228971454 0.077604381
Trifprat 0.232524328 0.128553835 0.0437890155 -0.042611817 0.089272402
Trifrepe -0.322607335 0.422132017 0.1215698753 0.383096915 -0.067705602
Vicilath -0.352398143 0.040430177 0.0618260529 -0.177126252 -0.118331138
Bracruta 0.159976508 0.205823892 -0.0964632149 0.102689199 -0.085332107
Callcusp 0.018570846 0.167200631 -0.0181969123 0.034626421 0.067508897
PC11 PC12 PC13 PC14 PC15
Achimill 0.132717925 0.134230514 -0.034529992 0.216182453 -0.141401817
Agrostol -0.059618860 -0.073683574 -0.023668258 -0.123235118 -0.106634349
Airaprae -0.019070127 -0.058358107 0.012396331 0.006629187 0.077238090
Alopgeni 0.039952489 -0.472711933 -0.065874698 0.056133072 0.142673465
Anthodor 0.099085859 0.039405527 0.030439268 -0.182295662 -0.191244567
Bellpere 0.191229646 -0.339813778 0.178784646 -0.145988631 -0.020153703
Bromhord 0.109494416 0.244018820 0.037703343 0.079868176 -0.018373727
Chenalbu -0.227060023 0.018287239 0.171677291 -0.175868511 -0.279195670
Cirsarve -0.003959815 0.141735155 0.001321334 0.049219524 0.036375284
Comapalu -0.051236512 0.049521631 0.419309406 0.122054623 -0.083871507
Eleopalu -0.131378144 -0.080553382 0.131165765 -0.207537309 0.007388714
Elymrepe 0.007174943 0.106204727 -0.030839977 -0.418387219 -0.195258305
Empenigr -0.304035391 -0.042476542 -0.027675462 -0.013779008 -0.351380876
Hyporadi -0.161835620 0.006085387 -0.045001999 -0.226512749 0.026476252
Juncarti -0.135408677 0.460006428 0.263053866 -0.143056615 -0.011814909
Juncbufo 0.331943950 0.383004314 -0.131204452 0.096142350 -0.059442194
Lolipere -0.399550594 0.009446008 -0.192536364 0.230441982 -0.122499231
Planlanc 0.112734237 0.045729471 0.043171048 -0.234799012 0.262590230
Poaprat -0.325080178 0.091586125 -0.011846325 0.019010072 0.047744134
Poatriv -0.209438657 -0.058924074 0.084029572 -0.135860829 0.013058855
Ranuflam -0.253468364 0.167283523 0.028297656 0.048388895 0.121361026
Rumeacet -0.017167126 0.032043329 -0.022347327 -0.179664647 0.114973393
Sagiproc 0.010890721 0.227060385 -0.066484762 0.028308217 0.124594706
Salirepe 0.042105059 0.100015813 -0.222850655 0.290392613 -0.071386194
Scorautu -0.181281016 0.048423771 0.228628199 -0.063119924 0.505549613
Trifprat -0.316843001 -0.089080821 -0.027087807 0.076188887 0.007168476
Trifrepe -0.198256965 0.071632853 -0.243172011 0.018039774 -0.180390197
Vicilath 0.148055454 0.120910330 -0.060677691 -0.428538696 -0.180520174
Bracruta 0.122374527 -0.190949281 0.161598801 0.073266522 -0.429555487
Callcusp -0.045372709 -0.035110432 -0.635088337 -0.283642351 0.111397731
PC16 PC17 PC18 PC19 PC20
Achimill 0.006319455 -0.250624910 0.130057211 -0.15259224 -0.280840132
Agrostol 0.129552488 0.220228162 0.032570554 -0.33376824 0.117562664
Airaprae 0.210143799 0.075340772 0.313245566 -0.17648404 -0.028419294
Alopgeni 0.141347045 0.067080980 -0.209999376 -0.17097107 0.077234388
Anthodor 0.389496778 0.221157627 -0.313756777 0.05751084 0.086145893
Bellpere 0.094710988 0.044306606 0.001176329 0.06773279 -0.233103744
Bromhord -0.374362864 0.041343305 -0.053691688 -0.14594158 0.305384469
Chenalbu 0.006875299 -0.091678747 0.173724870 0.09615198 -0.086869927
Cirsarve 0.143093017 0.062180079 0.221579806 0.00292687 0.004781978
Comapalu 0.024471933 0.243764018 -0.231073572 -0.23547941 -0.203407714
Eleopalu -0.316846051 0.038250547 0.153274202 0.26832548 -0.338905630
Elymrepe -0.003495925 -0.192734275 -0.038247140 -0.14099381 0.058019662
Empenigr -0.341052828 0.242487410 -0.163670390 0.27686627 0.252889870
Hyporadi -0.091888742 -0.083198813 0.020109210 -0.44286040 -0.372522205
Juncarti 0.235944145 0.009026855 0.102938841 0.02591653 0.001586611
Juncbufo -0.191629105 0.329604360 0.162314310 -0.18263770 -0.071387835
Lolipere -0.160510751 0.048846040 -0.219803850 -0.21507924 -0.173345126
Planlanc -0.056120436 0.405994596 -0.104807822 0.05804021 -0.114153392
Poaprat 0.213962048 0.415537482 0.145898300 0.13921121 -0.091198432
Poatriv -0.136171340 0.082075532 -0.069242868 -0.10994713 -0.207521475
Ranuflam 0.119804645 -0.138589298 -0.347714187 -0.20643369 0.135135821
Rumeacet -0.053875908 -0.155850682 -0.220366196 0.02634990 -0.124352068
Sagiproc 0.031989882 -0.272388398 -0.267648595 0.29545079 -0.279033917
Salirepe 0.121935233 0.114820087 -0.173474729 -0.02796627 -0.265406160
Scorautu -0.079587214 -0.030689931 0.151345426 -0.09463241 0.158732103
Trifprat 0.022440237 -0.167076686 0.220113547 -0.15845608 0.210115585
Trifrepe 0.366480663 -0.060442109 0.127395719 0.15596121 -0.023897121
Vicilath 0.030320179 -0.106893186 -0.142322601 -0.14990616 0.134383490
Bracruta -0.061532092 -0.010759652 0.187268280 -0.13283957 -0.052130439
Callcusp -0.085439050 0.140108877 0.131246343 -0.03698375 -0.057840003
> # plot(tmp)
> # barplot(log(tmp$sdev) ^ 2, names.arg = seq(1:4),
> # xlab = "component",
> # ylab = "variance")
>
> data(USArrests) # 1973年のアメリカにおける犯罪に関するデータ(犯罪の種類および都市部人口の割合、州)
> prcomp(~ Murder + Assault + Rape,
+ data = USArrests,
+ scale = TRUE)
Standard deviations:
[1] 1.5357670 0.6767949 0.4282154
Rotation:
PC1 PC2 PC3
Murder -0.5826006 0.5339532 -0.6127565
Assault -0.6079818 0.2140236 0.7645600
Rape -0.5393836 -0.8179779 -0.1999436
> # plot(prcomp(USArrests))
> summary(prcomp(USArrests, scale = TRUE))
Importance of components:
PC1 PC2 PC3 PC4
Standard deviation 1.5749 0.9949 0.59713 0.41645
Proportion of Variance 0.6201 0.2474 0.08914 0.04336
Cumulative Proportion 0.6201 0.8675 0.95664 1.00000
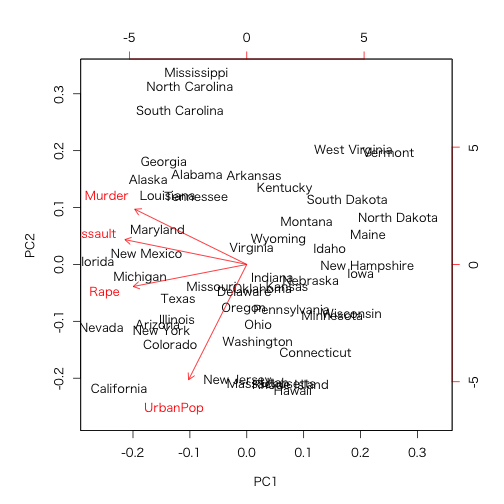
> biplot(prcomp(USArrests, scale = TRUE))
prop.table
割合テーブルの作成
> matrix(1:4, 2) %>% {
+ print(.)
+ prop.table(., 1)
+ }
[,1] [,2]
[1,] 1 3
[2,] 2 4
[,1] [,2]
[1,] 0.2500000 0.7500000
[2,] 0.3333333 0.6666667
terms
> ctl <- c(4.17,5.58,5.18,6.11,4.50,4.61,5.17,4.53,5.33,5.14)
> trt <- c(4.81,4.17,4.41,3.59,5.87,3.83,6.03,4.89,4.32,4.69)
> group <- gl(2, 10, 20, labels = c("Ctl","Trt"))
> weight <- c(ctl, trt)
>
> lm(weight ~ group) %>% terms()
weight ~ group
attr(,"variables")
list(weight, group)
attr(,"factors")
group
weight 0
group 1
attr(,"term.labels")
[1] "group"
attr(,"order")
[1] 1
attr(,"intercept")
[1] 1
attr(,"response")
[1] 1
attr(,".Environment")
<environment: 0x10c24bd20>
attr(,"predvars")
list(weight, group)
attr(,"dataClasses")
weight group
"numeric" "factor"
window
時系列データの抽出
Arguments
- x
- start
- end
- frequency, deltat
- extend
- ...
- value
> window(x = presidents, start = 1960, end = c(1969, 4))
Qtr1 Qtr2 Qtr3 Qtr4
1960 71 62 61 57
1961 72 83 71 78
1962 79 71 62 74
1963 76 64 62 57
1964 80 73 69 69
1965 71 64 69 62
1966 63 46 56 44
1967 44 52 38 46
1968 36 49 35 44
1969 59 65 65 56