ggtree: a phylogenetic tree viewer for different types of tree annotations
系統樹を描画するためのパッケージ
- Bioconductor: http://www.bioconductor.org/packages/release/bioc/html/ggtree.html
- GitHub: https://github.com/GuangchuangYu/ggtree
- vignettes
- viewing and annotating phylogenetic tree with ggtree
- updating a tree view using %<% operator
- an example of drawing beast tree using ggtree
- flip and rotate branches in ggtree
- subsetting data in ggtree
- ggtree with funny fonts
- comic phylogenetic tree with ggtree and comicR
- embeding a subplot in ggplot via subview
- annotate phylogenetic tree with local images
- phylomoji with ggtree
> library(ggtree)
Creating a generic function for 'nchar' from package 'base' in package 'S4Vectors'
Attaching package: 'ggtree'
The following object is masked from 'package:github':
get.tree
The following object is masked from 'package:igraph':
%>%
The following objects are masked from 'package:raster':
flip, mask, rotate
バージョン: 1.3.4
| 関数名 | 概略 |
|---|---|
%<% |
%<% |
%<+% |
%<+% |
%>% |
pipe |
. |
. |
Date2decimal |
Date2decimal |
NJ |
NJ |
StatHilight |
StatHilight |
add_colorbar |
add_colorbar |
add_legend |
add_legend |
aes |
creates a lists of unevaluated expressions |
annotation_clade |
annotation_clade |
annotation_clade2 |
annotation_clade2 |
annotation_image |
annotation_image |
apeBoot |
apeBoot |
apeBootstrap-class |
Class "apeBootstrap" This class stores ape bootstrapping analysis result |
as.binary |
as.binary |
as.data.frame.phylo |
as.data.frame |
beast-class |
Class "beast" This class stores information of beast output |
codeml-class |
Class "codeml" This class stores information of output from codeml |
codeml_mlc-class |
Class "codeml_mlc" This class stores information of mlc file frm codeml output |
collapse |
collapse |
decimal2Date |
decimal2Date |
download.phylopic |
download.phylopic |
expand |
expand |
flip |
flip |
fortify.phylo |
fortify |
geom_aline |
geom_aline |
geom_hilight |
geom_hilight |
geom_nodepoint |
geom_nodepoint |
geom_point2 |
geom_point2 |
geom_rootpoint |
geom_rootpoint |
geom_segment2 |
geom_segment2 |
geom_text |
text annotations |
geom_text2 |
geom_text2 |
geom_tiplab |
geom_tiplab |
geom_tippoint |
geom_tippoint |
geom_tree |
geom_tree |
get.fields |
get.fields method |
get.offspring.tip |
get.offspring.tip |
get.path |
get.path |
get.phylopic |
get.phylopic |
get.placements |
get.placements method |
get.subs |
get.subs method |
get.tipseq |
get.tipseq method |
get.tree |
get.tree method |
get.treeinfo |
get.treeinfo method |
get.treetext |
get.treetext method |
getNodeNum |
getNodeNum |
getRoot |
getRoot |
get_clade_position |
get_clade_position |
get_heatmap_column_position |
get_heatmap_column_position |
get_taxa_name |
get_taxa_name |
ggplotGrob |
generate a ggplot2 plot grob |
ggtree |
visualizing phylogenetic tree and heterogenous associated data based on grammar of graphics 'ggtree' provides functions for visualizing phylogenetic tree and its associated data in R. |
gheatmap |
gheatmap |
gplot |
gplot |
groupClade |
groupClade method |
groupOTU |
groupOTU method |
groupOTU.phylo |
groupOTU.phylo |
gzoom |
gzoom method |
gzoom.phylo |
gzoom |
hilight |
hilight |
hyphy-class |
Class "hyphy" This class stores information of HYPHY output |
jplace-class |
Class "jplace" This class stores information of jplace file. |
mask |
mask |
merge_tree |
merge_tree |
msaplot |
msaplot |
nhx-class |
Class "nhx" This class stores nhx tree |
paml_rst-class |
Class "paml_rst" This class stores information of rst file from PAML output |
phangorn-class |
Class "phangorn" This class stores ancestral sequences inferred from 'phangorn' |
phyPML |
treeAnno.pml |
phylopic |
phylopic |
plot |
plot method |
pmlToSeq |
pmlToSeq |
r8s-class |
Class "r8s" This class stores output info from r8s |
raxml-class |
Class "raxml" This class stores RAxML bootstrapping analysis result |
read.baseml |
read.baseml |
read.beast |
read.beast |
read.codeml |
read.codeml |
read.codeml_mlc |
read.codeml_mlc |
read.hyphy |
read.hyphy |
read.jplace |
read.jplace |
read.nhx |
read.nhx |
read.paml_rst |
read.paml_rst |
read.r8s |
read.r8s |
read.raxml |
read.raxml |
read.tree |
read newick tree |
reroot |
reroot method |
rm.singleton.newick |
rm.singleton.newick |
rotate |
rotate |
rtree |
generate random tree |
scaleClade |
scaleClade |
scale_color |
scale_color method |
scale_x_ggtree |
scale_x_ggtree |
show,raxml-method |
show method |
subview |
subview |
theme_transparent |
theme_transparent |
theme_tree |
theme_tree |
theme_tree2 |
theme_tree2 |
write.jplace |
write.jplace |
> library(ape)
Attaching package: 'ape'
The following object is masked from 'package:ggtree':
rotate
The following objects are masked from 'package:igraph':
edges, mst, ring
The following objects are masked from 'package:raster':
rotate, zoom
geom_tree
ggplot2用の系統樹レイヤー。ggplot()にはphyloクラスのデータを渡す
> rtree(n = 10) %$% ggplot(data = .) + geom_tree()
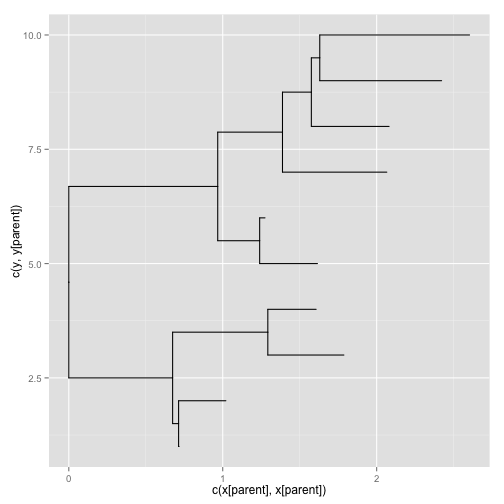
ggtree
ggplot2風に系統樹を描画
> rtree(10) %>% ggtree(.)
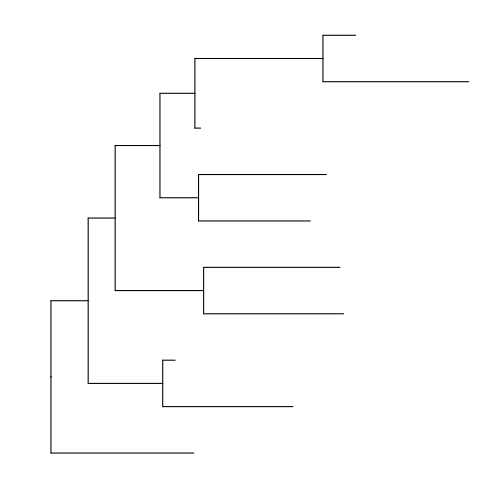
theme_tree
描画用テーマ
Arguments
- bgcolor
- fgcolor
> rtree(10) %>% ggtree(.) + theme_tree(bgcolor = "white", fgcolor = "black")
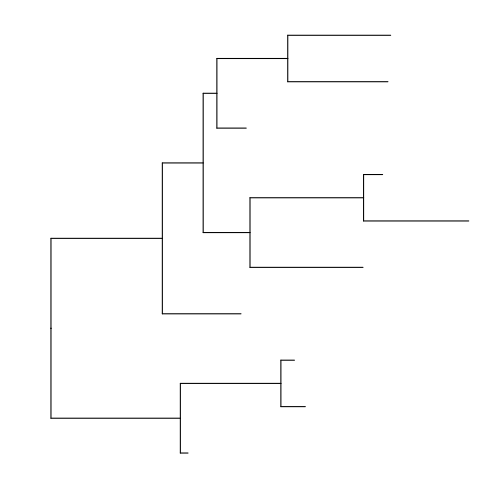
rtee
ランダムな系統樹データを作成する
ape::rtreeもある
Arguments
- n: 系統樹の先端となる葉の数
- rooted: logical
- tip.label: 葉に使用するラベル
- br
- ...
> rtree(n = 10, rooted = TRUE, tip.label = NULL, br = runif)
Phylogenetic tree with 10 tips and 9 internal nodes.
Tip labels:
t8, t10, t9, t2, t6, t3, ...
Rooted; includes branch lengths.