ggmcmc: Tools for Analyzing MCMC Simulations from Bayesian Inference
MCMCシュミレーション結果の解析ツール
- CRAN: http://cran.r-project.org/web/packages/ggmcmc/index.html
- GitHub: https://github.com/xfim/ggmcmc
- URL: http://xavier-fim.net/packages/ggmcmc
> library(ggmcmc)
Loading required package: dplyr
Attaching package: 'dplyr'
The following object is masked from 'package:sfsmisc':
last
The following object is masked from 'package:MASS':
select
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
Loading required package: tidyr
Attaching package: 'tidyr'
The following object is masked from 'package:coenocliner':
expand
The following object is masked from 'package:rstan':
extract
The following object is masked from 'package:Matrix':
expand
The following object is masked from 'package:magrittr':
extract
Loading required package: GGally
Warning: replacing previous import by 'grid::arrow' when loading 'GGally'
Warning: replacing previous import by 'grid::unit' when loading 'GGally'
Attaching package: 'GGally'
The following object is masked from 'package:dplyr':
nasa
> data("radon")
バージョン: 0.7.2
| 関数名 | 概略 |
|---|---|
ac |
Calculate the autocorrelation of a single chain, for a specified amount of lags |
calc_bin |
Calculate binwidths by parameter, based on the total number of bins. |
ci |
Calculate Credible Intervals (wide and narrow). |
get_family |
Subset a ggs object to get only the parameters with a given regular expression. |
ggmcmc |
Wrapper function that creates a single pdf file with all plots that ggmcmc can produce. |
ggs |
Import MCMC samples into a ggs object than can be used by all ggs_* graphical functions. |
ggs_Rhat |
Dotplot of Potential Scale Reduction Factor (Rhat) |
ggs_autocorrelation |
Plot an autocorrelation matrix |
ggs_caterpillar |
Caterpillar plot with thick and thin CI |
ggs_chain |
Auxiliary function that extracts information from a single chain. |
ggs_compare_partial |
Density plots comparing the distribution of the whole chain with only its last part. |
ggs_crosscorrelation |
Plot the Cross-correlation between-chains |
ggs_density |
Density plots of the chains |
ggs_geweke |
Dotplot of the Geweke diagnostic, the standard Z-score |
ggs_histogram |
Histograms of the paramters. |
ggs_pairs |
Create a plot matrix of posterior simulations |
ggs_ppmean |
Posterior predictive plot comparing the outcome mean vs the distribution of the predicted posterior means. |
ggs_ppsd |
Posterior predictive plot comparing the outcome standard deviation vs the distribution of the predicted posterior standard deviations. |
ggs_rocplot |
Receiver-Operator Characteristic (ROC) plot for models with binary outcomes |
ggs_running |
Running means of the chains |
ggs_separation |
Separation plot for models with binary response variables |
ggs_traceplot |
Traceplot of the chains |
gl_unq |
Generate a factor with unequal number of repetitions. |
radon |
Simulations of the parameters of a hierarchical model using the radon example in Gelman & Hill (2007). |
roc_calc |
Calculate the ROC curve for a set of observed outcomes and predicted probabilities |
s |
Simulations of the parameters of a simple linear regression with fake data. |
s.binary |
Simulations of the parameters of a simple linear regression with fake data. |
s.y.rep |
Simulations of the posterior predictive distribution of a simple linear regression with fake data. |
sde0f |
Spectral Density Estimate at Zero Frequency. |
y |
Values for the observed outcome of a simple linear regression with fake data. |
y.binary |
Values for the observed outcome of a binary logistic regression with fake data. |
ac
自己相関の検証。ggs_autocorretation()で用いられている。
> ac(cumsum(rnorm(10)) / 10, nLags = 5)
Lag Autocorrelation
1 1 1.0000000
2 2 0.8713890
3 3 0.7159419
4 4 0.8814559
5 5 0.9853668
ci
信用区間の算出
> s %>% ggs() %>% ci() %>%
+ kable()
Error in eval(expr, envir, enclos): object 's' not found
ggmcmc
{ggmcmc}で描画できるすべてのプロットをPDFとして保存
Arguments
- D
- file... 保存するPDFファイル名。初期値は"ggmcmc-output.pdf"
- family
- plot... 描画対象にする関数。文字列として関数名を与える。
- param_page
- width
- height
- simplify_traceplot
- ...
> s %>% ggs() %>%
+ ggmcmc()
> s %>% ggs() %>%
+ ggmcmc(plot = c("ggs_running", "ggs_autocorrelation"))
ggs
MCMC事後サンプルを{ggmcmc}内のggs_*()で利用可能なggsオブジェクト(data.frame, tbl_dfクラス)へ変換。反復回数、チェインの数、パラメータ名、値の列を持つ。
Arguments
- S... samples
- family
- description
- burnin
- par_labels
- inc_warmup
- stan_include_auxiliar
> s %>% ggs() %>% {
+ print(class(.))
+ dplyr::glimpse(.)
+ }
Error in eval(expr, envir, enclos): object 's' not found
> radon$s.radon.short %>% ggs() %>% {
+ print(class(.))
+ dplyr::glimpse(.)
+ }
[1] "tbl_df" "tbl" "data.frame"
Observations: 16,000
Variables: 4
$ Iteration (int) 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 1...
$ Chain (int) 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1...
$ Parameter (fctr) alpha[1], alpha[1], alpha[1], alpha[1], alpha[1], a...
$ value (dbl) 1.9675268, 1.0572830, 1.1732732, 1.2637594, 1.036500...
ggs_autocorrelation
> data("linear")
> s %>% ggs() %>% ggs_autocorrelation()

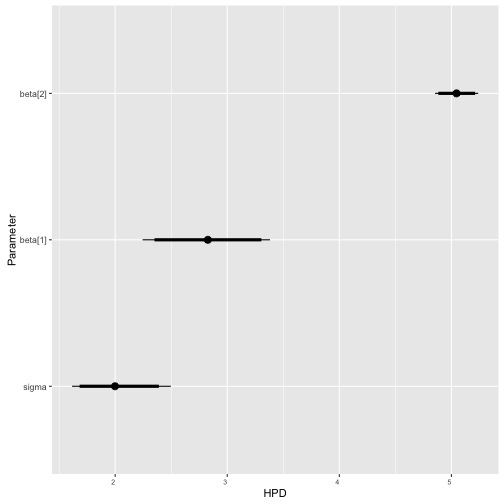
ggs_caterpillar
> data("linear")
> s %>% ggs() %>% ggs_caterpillar()
ggs_compare_partial
> data("linear")
> s %>% ggs() %>% ggs_compare_partial()

ggs_crosscorrelation
> data("linear")
> s %>% ggs() %>% ggs_crosscorrelation()

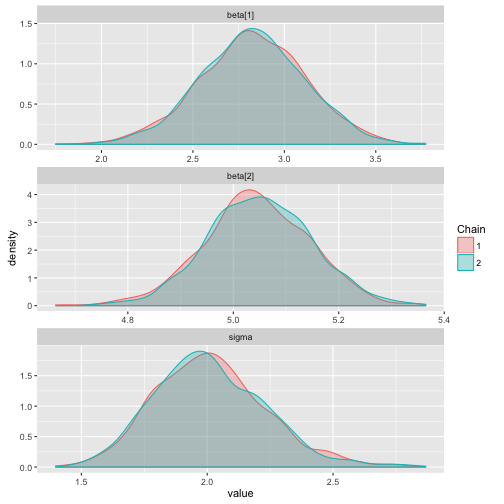
ggs_density
> data("linear")
> s %>% ggs() %>% ggs_density
ggs_geweke
> data("linear")
> s %>% ggs() %>% ggs_geweke()

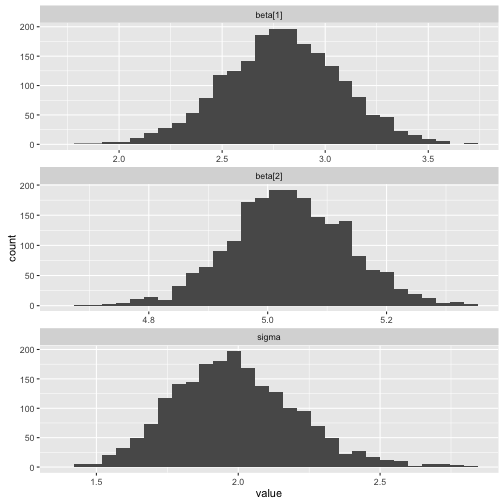
ggs_histogram
> data("linear")
> s %>% ggs() %>% ggs_histogram()
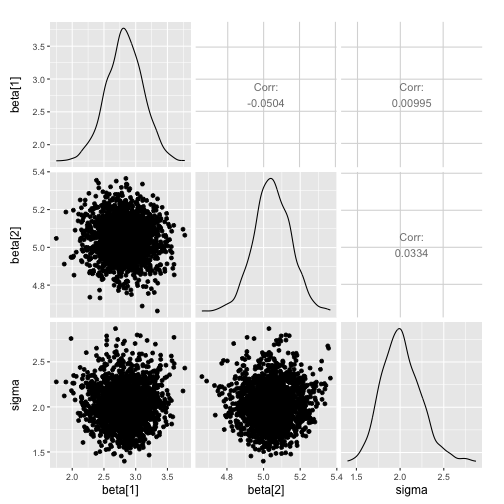
ggs_pairs
> data("linear")
> s %>% ggs() %>% ggs_pairs()
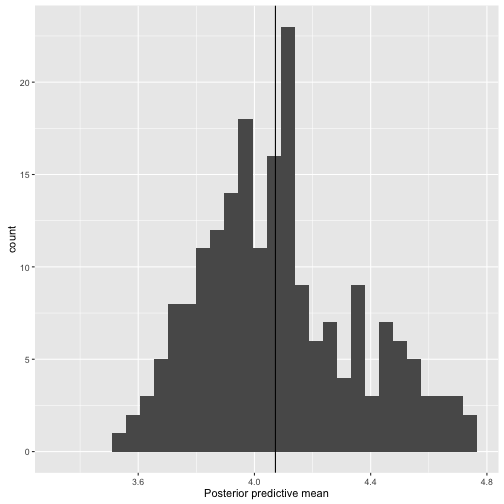
ggs_ppmean
> data("linear")
> s.y.rep %>% ggs() %>% ggs_ppmean(outcome = y)
Error in ggs_ppmean(., outcome = y): The length of the outcome must be equal to the number of Parameters of the ggs object.
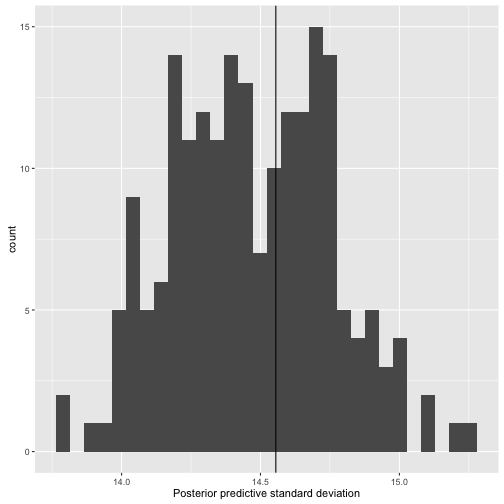
ggs_ppsd
> data("linear")
> s.y.rep %>% ggs() %>% ggs_ppsd(outcome = y)
Error in ggs_ppsd(., outcome = y): The length of the outcome must be equal to the number of Parameters of the ggs object.
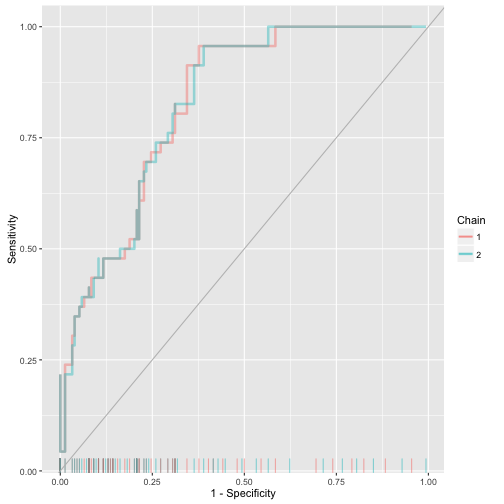
ggs_rocplot
> data("binary")
> s.binary %>% ggs(family = "mu") %>% ggs_rocplot(outcome = y.binary)

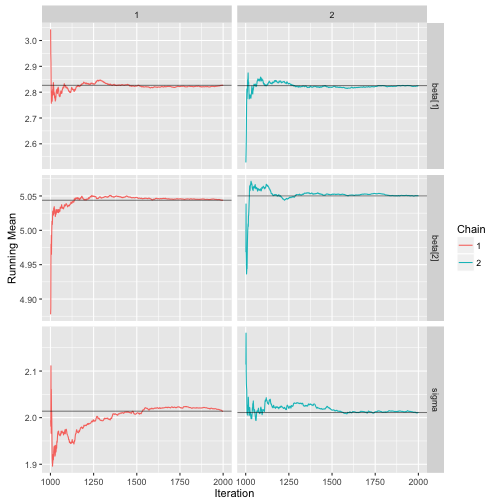
ggs_running
> data("linear")
> s %>% ggs() %>% ggs_running()

ggs_separation
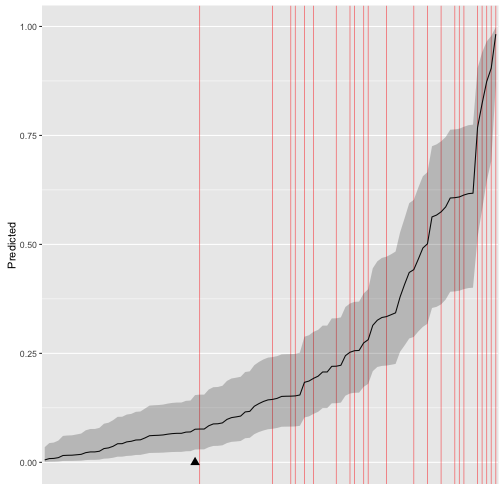
> data("binary")
> s.binary %>% ggs(family = "mu") %>% ggs_separation(outcome = y.binary)
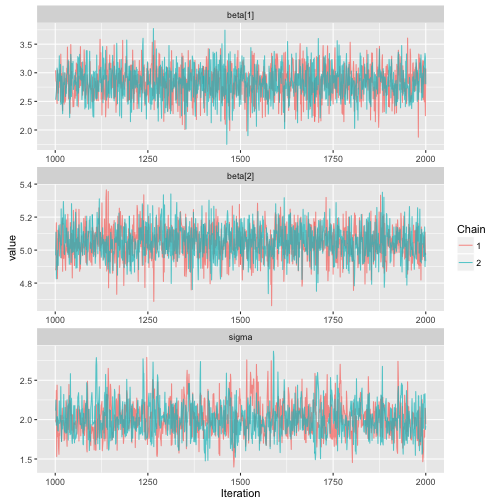
ggs_traceplot
> data("linear")
> s %>% ggs() %>% ggs_traceplot()
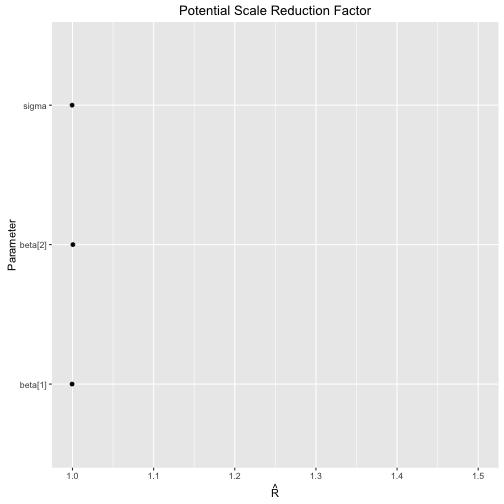
ggs_Rhat
> s %>% ggs() %>%
+ ggs_Rhat()
radon
> data("radon")
> radon %>% {
+ print(class(.))
+ print(names(.))
+ str(., max.level = 2)
+ }
[1] "list"
[1] "counties" "id.county" "y" "s.radon"
[5] "s.radon.yhat" "s.radon.short"
List of 6
$ counties :'data.frame': 85 obs. of 4 variables:
..$ County : chr [1:85] "Aitkin" "Anoka" "Becker" "Beltrami" ...
..$ id.county : num [1:85] 1 2 3 4 5 6 7 8 9 10 ...
..$ uranium : num [1:85] 0.502 0.429 0.893 0.552 0.867 ...
..$ ns.location: chr [1:85] "North" "South" "North" "North" ...
$ id.county : num [1:919] 1 1 1 1 2 2 2 2 2 2 ...
$ y : num [1:919] 0.788 0.788 1.065 0 1.131 ...
$ s.radon :List of 2
..$ : mcmc [1:100, 1:175] 1.186 1.119 1.034 0.969 1.282 ...
.. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "mcpar")= num [1:3] 10020 12000 20
..$ : mcmc [1:100, 1:175] 1.399 0.844 1.044 1.014 1.162 ...
.. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "mcpar")= num [1:3] 10020 12000 20
..- attr(*, "class")= chr "mcmc.list"
$ s.radon.yhat :List of 2
..$ : mcmc [1:10, 1:919] 0.807 0.597 0.111 0.374 0.29 ...
.. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "mcpar")= num [1:3] 32100 33000 100
..$ : mcmc [1:10, 1:919] 0.78984 0.32841 0.35366 -0.00246 0.39972 ...
.. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "mcpar")= num [1:3] 32100 33000 100
..- attr(*, "class")= chr "mcmc.list"
$ s.radon.short:List of 2
..$ : mcmc [1:2000, 1:4] 1.97 1.06 1.17 1.26 1.04 ...
.. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "mcpar")= num [1:3] 12010 32000 10
..$ : mcmc [1:2000, 1:4] 0.929 1.521 1.441 1.164 1.238 ...
.. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "mcpar")= num [1:3] 12010 32000 10
..- attr(*, "class")= chr "mcmc.list"
s
模擬データによる単回帰を行った推定結果。mcmc.listクラスオブジェクト
> s %>% {
+ print(class(.))
+ str(., max.level = 2)
+ }
[1] "mcmc.list"
List of 2
$ : mcmc [1:1000, 1:3] 3.04 2.85 2.85 2.58 2.46 ...
..- attr(*, "dimnames")=List of 2
..- attr(*, "mcpar")= num [1:3] 1001 2000 1
$ : mcmc [1:1000, 1:3] 2.53 2.59 2.67 2.81 3.08 ...
..- attr(*, "dimnames")=List of 2
..- attr(*, "mcpar")= num [1:3] 1001 2000 1
- attr(*, "class")= chr "mcmc.list"
s.binary
> s.binary %>% {
+ print(class(.))
+ str(., max.level = 2)
+ }
[1] "mcmc.list"
List of 2
$ : mcmc [1:100, 1:102] 0.01361 0.01663 0.00308 0.00167 0.06182 ...
..- attr(*, "dimnames")=List of 2
..- attr(*, "mcpar")= num [1:3] 1010 2000 10
$ : mcmc [1:100, 1:102] 0.04958 0.00532 0.01521 0.00817 0.00462 ...
..- attr(*, "dimnames")=List of 2
..- attr(*, "mcpar")= num [1:3] 1010 2000 10
- attr(*, "class")= chr "mcmc.list"
s.y.rep
> s.y.rep %>% {
+ print(class(.))
+ str(., max.level = 2)
+ }
[1] "mcmc.list"
List of 2
$ : mcmc [1:200, 1:50] -20.8 -21.9 -23.1 -19.3 -21.5 ...
..- attr(*, "dimnames")=List of 2
..- attr(*, "mcpar")= num [1:3] 2201 2400 1
$ : mcmc [1:200, 1:50] -25.1 -18.9 -22.3 -21 -17.9 ...
..- attr(*, "dimnames")=List of 2
..- attr(*, "mcpar")= num [1:3] 2201 2400 1
- attr(*, "class")= chr "mcmc.list"