downscale: Downscaling Species Occupancy
- CRAN: http://cran.r-project.org/web/packages/downscale/index.html
- Vignettes:
> library(downscale)
> data.file <- system.file("extdata", "atlas_data.txt", package = "downscale")
> atlas.data <- read.table(data.file, header = TRUE)
バージョン: 1.2.3
| 関数名 | 概略 |
|---|---|
downscale |
Model area of occupancy against grain size for downscaling |
downscale-package |
Downscaling Species Occupancy |
ensemble.downscale |
Ensemble modelling of multiple downscaling functions |
hui.downscale |
Predict occupancy at fine grain sizes using the Hui model |
plot.predict.downscale |
Plotting of downscaled occupancy at fine grain sizes |
predict.downscale |
Predict occupancy at fine grain sizes |
upgrain |
Upgraining of atlas data to larger grain sizes |
upgrain.threshold |
Exploration of trade-offs in threshold selection for upgraining |
downscale
Arguments
- occupancies
- model...
Nachman,PL,Logis,Poisson,NB,GNB,INB,FNB,Thomas - extent
- tolerance
- starting_params
> tail(atlas.data)
>
> # thresh <- upgrain.threshold(atlas.data = atlas.data,
> # cell.width = 10,
> # scales = 3,
> # thresholds = seq(0, 1, 0.1))
>
> occupancy <- upgrain(atlas.data,
+ cell.width = 10,
+ scales = 3,
+ plot = FALSE,
+ method = "All_Sampled")
>
> ## Logistic model
> (logis <- downscale(occupancies = occupancy,
+ model = "Logis"))
>
> ### predict occupancy at finer grain sizes
> pred <- predict(logis,
+ new.areas = c(1, 2, 5, 25, 100, 400, 1600, 6400))
>
> ### Plot predictions
> plot(pred)
ensemble.downscale
> ## upgrain data (using All Samples threshold)
> occupancy <- upgrain(atlas.data,
+ cell.width = 10,
+ scales = 3,
+ method = "All_Sampled",
+ plot = FALSE)
>
> ## ensemble downscaling with an object of class upgrain
> ensemble.downscale(occupancies = occupancy,
+ new.areas = c(1, 2, 5, 15, 50, 100, 400, 1600, 6400),
+ cell.width = 10,
+ models = c("Nachman", "PL", "Logis", "GNB", "FNB", "Hui"),
+ plot = FALSE)
Nachman model is running... complete
PL model is running... complete
Logis model is running... complete
GNB model is running... complete
FNB model is running... complete
Hui model is running... complete
$Occupancy
Cell.area Nachman PL Logis GNB FNB
1 1 0.08030822 0.1052773 0.05985936 0.08028227 0.008908909
2 2 0.09736358 0.1219021 0.07665411 0.09733771 0.017281943
3 5 0.12519339 0.1479747 0.10546243 0.12516905 0.039715181
4 15 0.16820070 0.1866865 0.15220476 0.16818092 0.095193389
5 50 0.23008459 0.2408337 0.22156981 0.23007347 0.196529689
6 100 0.27380698 0.2788650 0.27067303 0.27380220 0.265954200
7 400 0.38060345 0.3738932 0.38685570 0.38061083 0.403406870
8 1600 0.51186710 0.5013040 0.51752119 0.51187602 0.522672643
9 6400 0.65827020 0.6721323 0.64583371 0.65825516 0.621741960
Hui Means
1 0.1635464 0.06242884
2 0.1681488 0.07975388
3 0.1774062 0.10949177
4 0.1963087 0.15707798
5 0.2347679 0.22516875
6 NA 0.27258720
7 NA 0.38494506
8 NA 0.51299864
9 NA 0.65102374
$AOO
Cell.area Nachman PL Logis GNB FNB Hui
1 1 30838.36 40426.47 22985.99 30828.39 3421.021 62801.81
2 2 37387.62 46810.42 29435.18 37377.68 6636.266 64569.15
3 5 48074.26 56822.29 40497.57 48064.91 15250.630 68123.98
4 15 64589.07 71687.60 58446.63 64581.47 36554.262 75382.54
5 50 88352.48 92480.13 85082.81 88348.21 75467.401 90150.86
6 100 105141.88 107084.16 103938.44 105140.05 102126.413 NA
7 400 146151.73 143575.00 148552.59 146154.56 154908.238 NA
8 1600 196556.97 192500.74 198728.14 196560.39 200706.295 NA
9 6400 252775.76 258098.80 248000.14 252769.98 238748.913 NA
Means
1 23972.68
2 30625.49
3 42044.84
4 60317.95
5 86464.80
6 104673.49
7 147818.90
8 196991.48
9 249993.12
hui.downscale
> # downscale とpredict.downscaleの組み合わせ
> (hui <- hui.downscale(atlas.data,
+ cell.width = 10,
+ extent = 228900,
+ new.areas = c(1, 2, 5, 15, 50,75),
+ plot = FALSE))
$model
[1] "Hui"
$predicted
Cell.area Occupancy AOO
1 1 0.2787162 63798.13
2 2 0.2866964 65624.80
3 5 0.3026427 69274.90
4 15 0.3347401 76622.02
5 50 0.3979450 91089.62
6 75 0.4291749 98238.12
$observed
Cell.area Occupancy
1 100 0.4552206
attr(,"class")
[1] "predict.downscale"
predict.downscale
Arguments
- object
- new.areas
- tolerance
- plot
- ...
upgrain
Arguments
- atlas.data
- cell.width
- scales
- threshold
- method...
All_Sampled,All_Occurrences,Gain_Equals_LossorSampled_Only - plot
- return.rasters
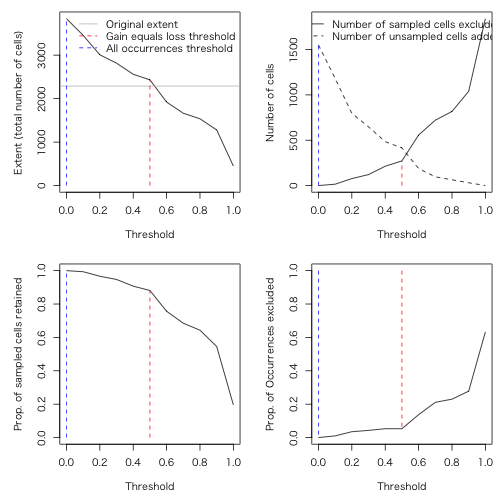
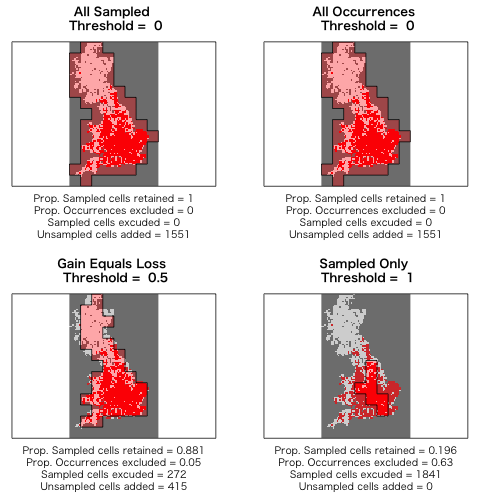
> thresh <- upgrain.threshold(atlas.data = atlas.data,
+ cell.width = 10,
+ scales = 3,
+ thresholds = seq(0, 1, 0.1))
> ## use a specified threshold - method must equal NULL
> upgrain(atlas.data = atlas.data,
+ cell.width= 10,
+ scales = 3,
+ threshold = 0.15,
+ plot = FALSE,
+ method = NULL)
$threshold
[1] 0.15
$extent.stand
[1] 320000
$occupancy.stand
Cell.area Extent Occupancy
1 100 320000 0.320
2 400 320000 0.455
3 1600 320000 0.555
4 6400 320000 0.700
$occupancy.orig
Cell.area Extent Occupancy
1 100 228900 0.4552206
2 400 266400 0.5645646
3 1600 312000 0.6102564
4 6400 384000 0.6666667
$atlas.raster.stand
class : RasterLayer
dimensions : 104, 64, 6656 (nrow, ncol, ncell)
resolution : 10, 10 (x, y)
extent : 8085, 8725, -65, 975 (xmin, xmax, ymin, ymax)
coord. ref. : NA
data source : in memory
names : layer
values : 1, 3 (min, max)
attr(,"class")
[1] "upgrain"